GUI Front-end of Data Processing Pipeline¶
Preprocessing¶
A ‘Confirmation’ option is provided in each of the following preprocessing step. When confirmation is required, the program displays the processing result after the target file is generated to ensure the correctness the final result.
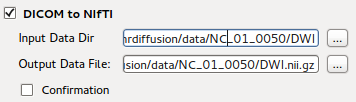
DICOM to NIFTI Conversion¶
This is the GUI front end for dcm2nii program. Details can be found at
DICOM to NIFTI Conversion section.
Eddy Current Correction¶
This is the GUI front end for bneddy program. Details can be found at
Data correction section.
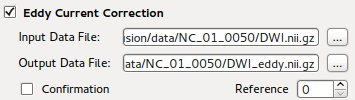
Since this is a time-consuming step in preprocessing stage, one can skip this step ignoring potential image registration error in original DWI data by unchecking the option. Then the converted NIFTI image would be input of brain extraction tool directly if the skull stripping option is enabled.
Skull Stripping¶
FSL’s BET (Brain Extraction Tool) is integrated as part of the program, so that one can verify automatic skull stripping result by visualizing overlaying mask image upon original image. See Volume Image Overlay on how to generate overlaid background images.
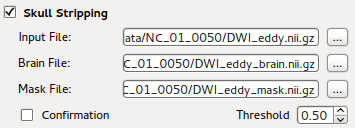
A threshold value that indicate ‘fractional intensity’ is exposed on the GUI, so that one can adjust the threshold to get smaller/larger brain outline estimates. Details on the implement can be found at BET’s UserGuide.
Diffusion Model Reconstruction¶
Starting from this section, we provide three different approaches for diffusion MR data processing:
This is the GUI front end for Reconstruction of the diffusion model, which generates required model for Fiber tracking and attributes extraction from DWI image and related b-values.
Click one of the options above to see guidelines on using the specific method for data processing.
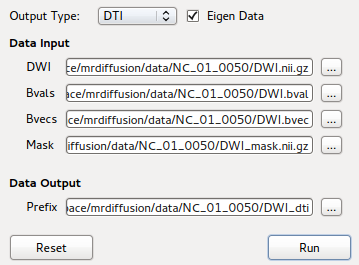
GUI front-end for DTI Reconstruction using bndti_estimate
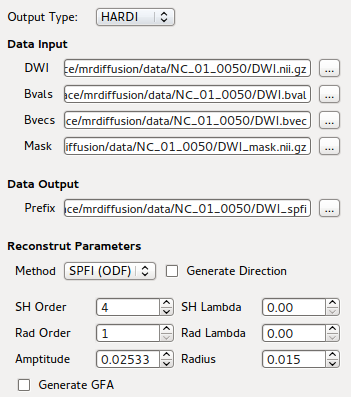
GUI front-end for SPFI Reconstruction using bnhardi_ODF_estimate
GUI front-end for CSD Reconstruction using bnhardi_FOD_estimate
Once DWI data is selected through file selection dialog, all other input and output options would be automatically generated. This simplifies the usage of command-line program, which requires a list of options to be entered.
Fiber Tracking¶
To generate fiber tracking result using previously generated diffusion model,
we provide the GUI front-end for using bndti_tracking and bnhardi_tracking,
which generate tract files from DTI and HARDI respectively.
The details of the program are described on Fiber tracking and attributes extraction section.
Fiber tracking require seed(ROI) files to initialize a tracking process.
One can either generate the seed files from the Registration Tool we provided from
Tools menu, or split a brain atlas image (e.g. AAL) using bncalc or bnroisplit
into multiple ROI files.
To generate whole brain fiber, just use the brain mask image generated during Skull Stripping
as seed input. This may cost a few minutes when HARDI is used as input diffusion model.
Miscellaneous Tools¶
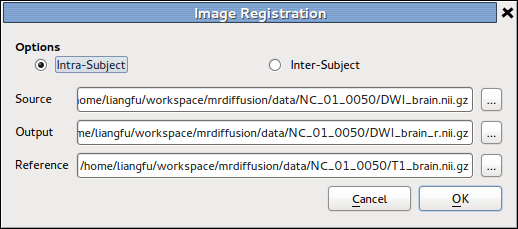
Image Registration¶
This is basically the GUI front-end of NiftyReg, while we perform reg_aladin
on intra-subject images, and addtionally run reg_f3d for inter-subject cases.
The details of the tool is described in Image Registration section in UserGuide page.
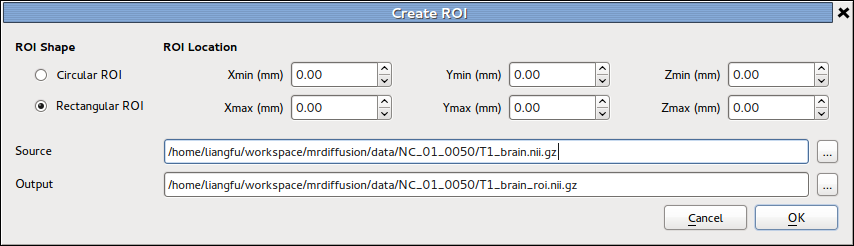
Create ROI¶
The details of the tool is described in Image calculation and ROI generation section in UserGuide page.
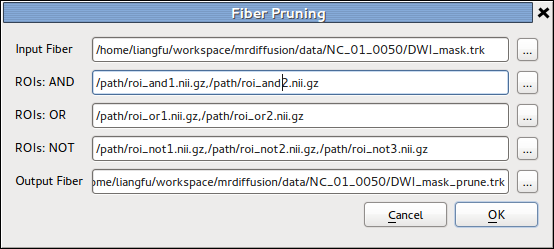
Fiber Pruning¶
The details of the tool is described in Fiber manipulation section in UserGuide page.