Overview¶
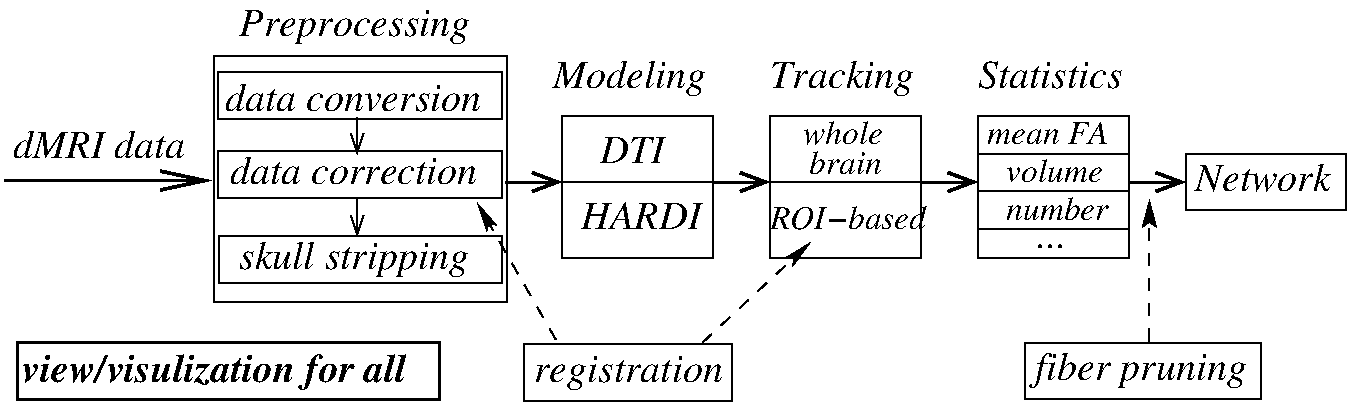
Brainnetome DiffusionKit is a light one-stop cross-platform solution to dMRI data analysis. The package delivers a complete pipeline from data format conversion to preprocessing, from local reconstruction to fiber tracking, and from fiber statistics to visualization. DiffusionKit was developed as a cross-platform framework, using ITK [1] for computation, VTK [2] for visualization, and Qt for GUI design. Both GPU and CPU computing were implemented for visualization to achieve high frame-rate, for rendering complex scene like whole brain tractographs in particular. The project was managed using the compiler-independent CMake [3], which is compatible with gcc/g++ and MS Visual Studio, etc. Well-established algorithms, such as the DICOM conversion tool dcm2nii by Chris Rorden [4] and the constrained spherical deconvolution (CSD) for HARDI reconstruction in MRtrix [5], were adopted with improved interface and user experience.
Key functions of the software¶
| Functions | Program | Description (use ‘-h’ argument for more details) |
|---|---|---|
| Preprocess | dcm2nii | Convert DICOM to unified 4D NIFTI files |
| bneddy | Correct eddy current induced distortion and head motion | |
| topup/eddy | Correct eddy current and susceptibility induced distortion and head movements | |
| bet2 | Extract brain tissue (Smith, 2002) | |
| bnsplit,bnmerge | Split/merge the 4D image along the 4th dimension | |
| Modeling | bndti_estimate | Estimate tensor model, output FA, MD, tensor etc. |
| bnhardi_ODF_estimate | Estimate ODF by SPFI method | |
| bnhardi_FOD_estimate | Estimate FOD by CSD method (Tournier et al., 2007) | |
| Tracking | bndti_tracking | Track white matter fiber based on tensor model |
| bnhardi_tracking | Track white matter fiber based on ODF/FOD | |
| Visualize | bnviewer | Visualize various kinds of data (.nii.gz, .trk) |
| Tools | bncalc,bnroisplit | Numeric calculation and ROI generation |
| bninfo | Show the head information of DICOM and NIFTI files | |
| reg_aladin,reg_f3d | Inter/intra-image registration across modalities | |
| reg_resample, reg_transform | Resample and apply transformation matrix | |
| bnfiber_end, bnfiber_prune | Prune fiber bundle, logical and/or/not based on ROIs | |
| bnfiber_stats | Export attributes of fiber bundles | |
| bnfiber_map | Generate the fiber density going through each voxel | |
| bnnetwork | Function to construct anatomical network |
It should be noteworthy that, for all the computing steps provided in GUI, the called commands with the complete parameter list are displayed in a separate log window. Such a design is special for the users to keep in mind what he/she is doing, and furthermore, it could be directly copied into script (Bash, Python …) for batch processing.